Note
Click here to download the full example code
Energy surface¶
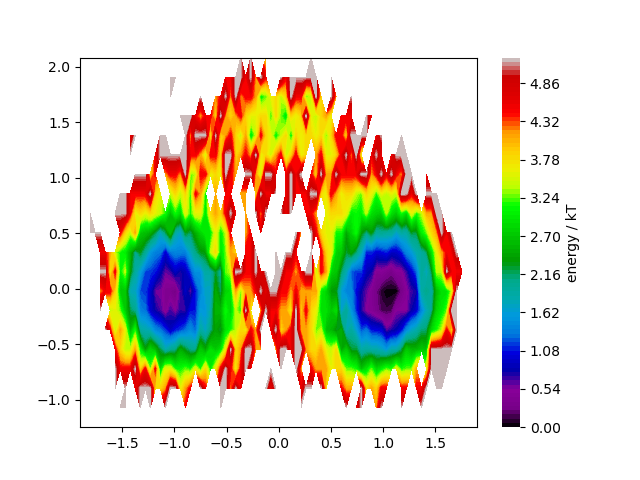
We show how to plot a two-dimensional energy surface based on util.energy2d
and plots.plot_energy2d.
8 import numpy as np
9
10 from deeptime.data import triple_well_2d
11 from deeptime.clustering import KMeans
12 from deeptime.markov.msm import MaximumLikelihoodMSM
13 from deeptime.util import energy2d
14
15 trajs = triple_well_2d(h=1e-3, n_steps=100).trajectory(x0=[[-1, 0], [1, 0], [0, 0]], length=5000)
16 traj_concat = np.concatenate(trajs, axis=0)
17 clustering = KMeans(n_clusters=20).fit_fetch(traj_concat)
18 dtrajs = [clustering.transform(traj) for traj in trajs]
19 msm = MaximumLikelihoodMSM(lagtime=1).fit_fetch(dtrajs)
20 weights = msm.compute_trajectory_weights(np.concatenate(dtrajs))[0]
21
22 energies = energy2d(*traj_concat.T, bins=(80, 20), kbt=1, weights=weights, shift_energy=True)
23 ax, contour, cbar = energies.plot(contourf_kws=dict(cmap='nipy_spectral'))
24 cbar.set_label('energy / kT')
Total running time of the script: ( 0 minutes 0.847 seconds)
Estimated memory usage: 9 MB
