Note
Click here to download the full example code
Chapman-Kolmogorov test¶
Tests the Chapman-Kolmogorov equality, see ck_test.
We demonstrate how to re-use the view grid to overlay two CK tests.
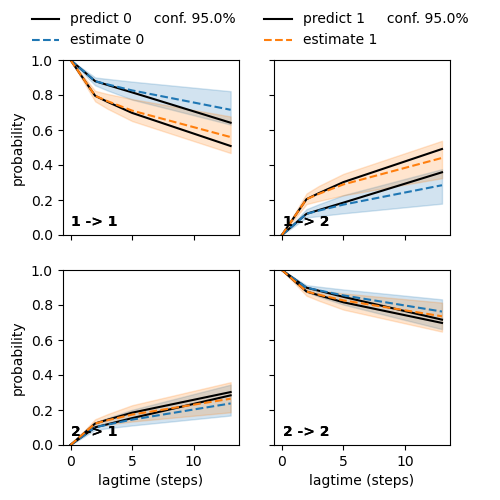
/home/mho/Development/deeptime/deeptime/plots/chapman_kolmogorov.py:174: ComplexWarning: Your CKtest contains imaginary components which are ignored during plotting.
warnings.warn("Your CKtest contains imaginary components which are ignored during plotting.",
9 from deeptime.clustering import KMeans
10 from deeptime.data import ellipsoids
11 from deeptime.markov import TransitionCountEstimator
12 from deeptime.markov.msm import BayesianMSM
13 from deeptime.plots.chapman_kolmogorov import plot_ck_test
14
15 traj = ellipsoids().observations(500)
16 traj2 = ellipsoids().observations(500)
17 dtraj = KMeans(n_clusters=15).fit_transform(traj)
18 dtraj2 = KMeans(n_clusters=15).fit_transform(traj2)
19
20 models = []
21 models2 = []
22 for lag in [2, 3, 5, 13]:
23 counts_estimator = TransitionCountEstimator(lagtime=lag, count_mode='effective')
24 models.append(BayesianMSM().fit_fetch(counts_estimator.fit_fetch(dtraj).submodel_largest()))
25 models2.append(BayesianMSM().fit_fetch(counts_estimator.fit_fetch(dtraj2).submodel_largest()))
26
27 test_model = models[0]
28 ck_test = test_model.ck_test(models, n_metastable_sets=2)
29 grid = plot_ck_test(ck_test, legend=False)
30
31 test_model2 = models2[0]
32 ck_test = test_model2.ck_test(models2, n_metastable_sets=2)
33 plot_ck_test(ck_test, legend=True, grid=grid)
Total running time of the script: ( 0 minutes 2.836 seconds)
Estimated memory usage: 11 MB
