function double_well_2d¶
- deeptime.data.double_well_2d(h=0.001, n_steps=10000, temperature_factor=1.0, mass=1.0, damping=1.0)¶
This dataset generates trajectories of a two-dimensional particle living in a double-well potential landscape.
The particle is subject to the stochastic differential equation
\[\mathrm{d}X_t = \nabla V(X_t) \mathrm{d}t + \sigma(t, X_t)\mathrm{d}W_t\]with \(W_t\) being a Wiener process, \(\sigma = \sqrt{\frac{\mathrm{kT}}{2\cdot m\cdot d}}\), where \(m\) is the mass, \(d\) the damping factor, and the potential \(V\) being given by
\[V(x) = (x_1^2 - 1)^2 + x_2^2.\](
Source code,png,hires.png,pdf)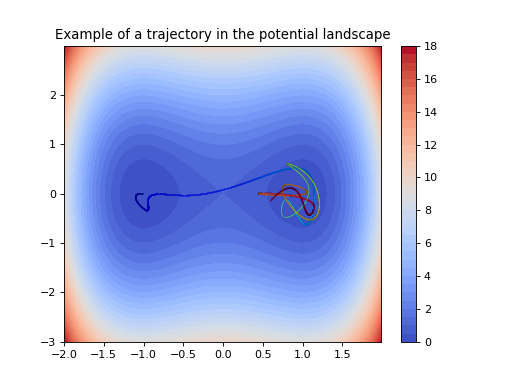
- Parameters:
h (float, default = 1e-3) – Integration step size. The implementation uses an Euler-Maruyama integrator.
n_steps (int, default = 10000) – Number of integration steps between each evaluation. That means the default lag time is
h*n_steps=0.1.temperature_factor (float, default=1) – The temperature kT.
mass (float, default=1) – The particle’s mass.
damping (float, default=1) – Damping factor.
- Returns:
model – The model.
- Return type:
Examples
The model possesses the capability to simulate trajectories as well as be evaluated at test points:
>>> import numpy as np >>> import deeptime as dt
First, set up the model (which internally already creates the integrator).
>>> model = dt.data.double_well_2d(h=1e-3, n_steps=100) # create model instance
Now, a trajectory can be generated:
>>> traj = model.trajectory(np.array([[-1., 0.]]), 1000, seed=42, n_jobs=1) # simulate trajectory >>> assert traj.shape == (1000, 2) # 1000 evaluations from initial condition [0, 0]
Or, alternatively the model can be evaluated at test points (mapping forward using the dynamical system):
>>> test_points = np.random.uniform(-2, 2, (100, 2)) # 100 test point in [-2, 2] x [-2, 2] >>> evaluations = model(test_points, seed=53, n_jobs=1) >>> assert evaluations.shape == (100, 2)
