Note
Click here to download the full example code
Triple-well 2D¶
Example for the deeptime.data.triple_well_2d() dataset.
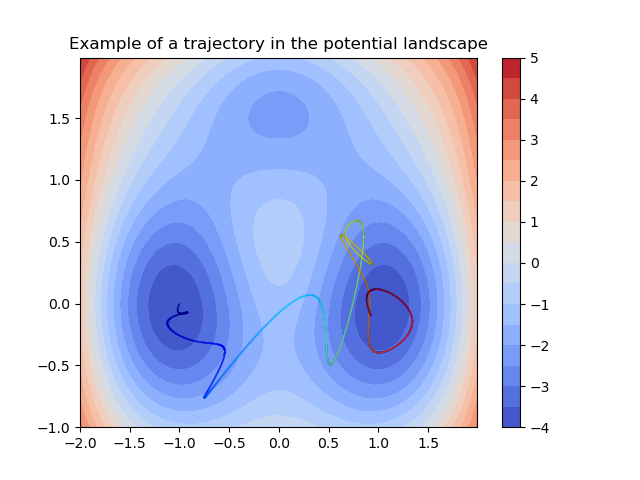
9 import numpy as np
10 import scipy
11 import matplotlib.pyplot as plt
12 from matplotlib.collections import LineCollection
13
14 from deeptime.data import triple_well_2d
15
16 system = triple_well_2d(n_steps=10000)
17 traj = system.trajectory(np.array([[-1, 0]]), 20, seed=42)
18
19 x = np.arange(-2, 2, 0.01)
20 y = np.arange(-1, 2, 0.01)
21 xy = np.meshgrid(x, y)
22 V = system.potential(np.dstack(xy).reshape(-1, 2)).reshape(xy[0].shape)
23
24 fig, ax = plt.subplots(1, 1)
25 ax.set_title("Example of a trajectory in the potential landscape")
26
27 cb = ax.contourf(x, y, V, levels=20, cmap='coolwarm')
28
29 x = np.r_[traj[:, 0]]
30 y = np.r_[traj[:, 1]]
31 f, u = scipy.interpolate.splprep([x, y], s=0, per=False)
32 xint, yint = scipy.interpolate.splev(np.linspace(0, 1, 50000), f)
33
34 points = np.stack([xint, yint]).T.reshape(-1, 1, 2)
35 segments = np.concatenate([points[:-1], points[1:]], axis=1)
36 coll = LineCollection(segments, cmap='jet')
37 coll.set_array(np.linspace(0, 1, num=len(points), endpoint=True))
38 coll.set_linewidth(1)
39 ax.add_collection(coll)
40
41 fig.colorbar(cb)
Total running time of the script: ( 0 minutes 0.783 seconds)
Estimated memory usage: 22 MB
