Kernel EDMD¶
This notebook illustrates how to use kernel extended dynamic mode decomposition (kernel EDMD) [1] to compute eigenfunctions of the Koopman operator and Perron-Frobenius operator.
An alternative derivation of kernel EDMD using covariance and cross-covariance operators defined on reproducing kernel Hilbert spaces, highlighting also relationships with conditional mean embeddings of distributions and the Perron-Frobenius operator, is presented in [2]. High-dimensional molecular dynamics problems can be found in [3].
Given training data \(\{x_i\}_{i=1}^m\) and \(\{y_i\}_{i=1}^m\), where \(y_i\) is a time-lagged version of \(x_i\), let \(\varphi\) denote an eigenfunction and \(\lambda\) the corresponding eigenvalue. For the Perron-Frobenius operator, we obtain kernel-based approximations of eigenvalues and eigenfunctions by
and for the Koopman operator by
where \(\Phi=[\phi(x_1), \dots, \phi(x_m)]\) is the so-called feature matrix and \(\varepsilon\) a regularization parameter. The matrices \(G_{XX}\) and \(G_{XY}\) are the standard Gram matrix and the time-lagged Gram matrix, respectively, with entries \([G_{XX}]_{ij} = k(x_i, x_j)\) and \([G_{XY}]_{ij} = k(x_i, y_j)\). Moreover, \(G_{YX}=G_{XY}^\top\).
[1]:
import numpy as np
import matplotlib.pyplot as plt
First we define a dynamical system. We will use a quadruple-well problem as our guiding example, which is given by a stochastic differential equation of the form
The potential \(V\) is defined by
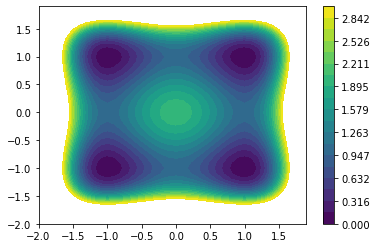
We define the inverse temperature to be \(\beta = 4\). Let us visualize the potential.
[2]:
xy = np.arange(-2, 2, 0.1)
XX, YY = np.meshgrid(xy, xy)
V = (XX**2 - 1)**2 + (YY**2 - 1)**2
plt.contourf(xy, xy, V, levels=np.linspace(0.0, 3.0, 20))
plt.colorbar();
The dynamical system itself is implented in C++ for the sake of efficiency. The SDE is solved with the Euler-Maruyama method.
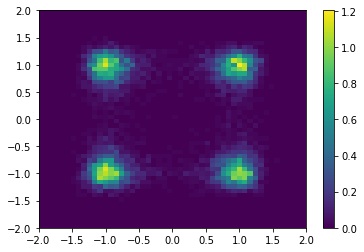
To visualize the system’s dynamics we generate one long trajectory and plot a histogram. The density is high where the potential is low and vice versa. A particle will typically stay in one well for a long time before it transitions to other wells. There are thus four metastable states corresponding to the four wells.
We can load the model as follows.
[3]:
from deeptime.data import quadruple_well
h = 1e-3 # step size of the Euler-Maruyama integrator
n_steps = 10000 # number of steps, the lag time is thus tau = nSteps*h = 10
x0 = np.zeros((1, 2)) # inital condition
n = 10000 # number of evaluations of the discretized dynamical system with lag time tau
f = quadruple_well(n_steps=n_steps) # loading the model
traj = f.trajectory(x0, n, seed=42)
plt.hist2d(*traj.T, range=[[-2, 2], [-2, 2]], bins=50, density=True)
plt.colorbar();
To approximate the Koopman operator, we uniformly sample training data in \([-2, 2] \times [-2, 2]\).
[4]:
m = 2500 # number of training data points
X = np.random.uniform(-2, 2, size=(2500, 2)) # training data
# X = 4*np.random.rand(2, m)-2
Y = f(X, seed=42, n_jobs=1) # training data mapped forward by the dynamical system
We define a Gaussian kernel with bandwidth \(\sigma=1\) and apply kernel EDMD to the data set.
[5]:
from deeptime.kernels import GaussianKernel
from deeptime.decomposition import KernelEDMD
sigma = 1 # kernel bandwidth
kernel = GaussianKernel(sigma)
n_eigs = 6 # number of eigenfunctions to be computed
kedmd_estimator = KernelEDMD(kernel, n_eigs=n_eigs, epsilon=1e-3)
kedmd_model = kedmd_estimator.fit((X, Y)).fetch_model()
phi = np.real(kedmd_model.transform(X))
# normalize
for i in range(n_eigs):
phi[:, i] = phi[:, i] / np.max(np.abs(phi[:, i]))
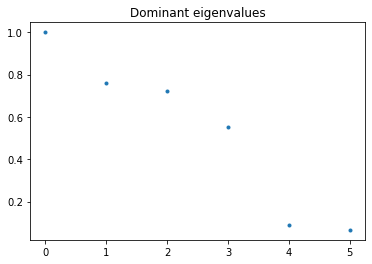
We expect four eigenvalues close to one. These dominant eigenvalues correspond to the slow dynamics. The eigenfunction of the Koopman operator associated with the eigenvalue \(\lambda=1\) is the constant function, and the eigenfunction of the Perron-Frobenius operator associated with this eigenvalue is the invariant density.
[6]:
d = np.real(kedmd_model.eigenvalues)
plt.plot(d, '.')
plt.title('Dominant eigenvalues');
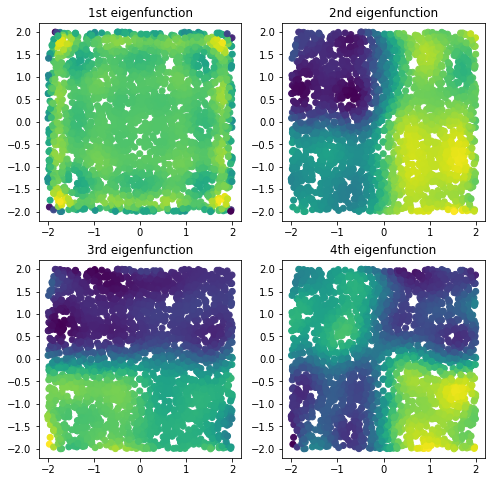
The eigenfunctions contain information about the metastable sets and slow transitions.
[7]:
fig = plt.figure(figsize=(8, 8))
gs = fig.add_gridspec(ncols=2, nrows=2)
ax = fig.add_subplot(gs[0, 0])
ax.scatter(*X.T, c=phi[:, 0])
ax.set_title('1st eigenfunction')
ax = fig.add_subplot(gs[0, 1])
ax.scatter(*X.T, c=phi[:, 1])
ax.set_title('2nd eigenfunction')
ax = fig.add_subplot(gs[1, 0])
ax.scatter(*X.T, c=phi[:, 2])
ax.set_title('3rd eigenfunction')
ax = fig.add_subplot(gs[1, 1])
ax.scatter(*X.T, c=phi[:, 3])
ax.set_title('4th eigenfunction');
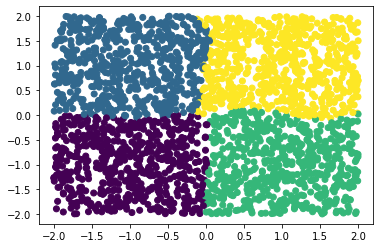
We can now apply clustering techniques to the dominant eigenfunctions to obtain metastable sets.
[8]:
from deeptime.clustering import KMeans
kmeans = KMeans(n_clusters=4, n_jobs=4).fit(phi[:, :4]).fetch_model()
c = kmeans.transform(phi[:, :4])
plt.scatter(*X.T, c=c);
References
