EDMD¶
For people already familiar with the method and interface, the API docs.
Similarly to DMD, extended DMD (EDMD) [1] is a data-driven method to approximate the Koopman operator. The Koopman operator \(\mathcal{K}\) describes how an observable of an (potentially stochastic) dynamical system evolves in time. In a discrete-time setting with \(x_{t+1} = T(x_t)\) and an observable \(g\), the Koopman operator is defined as
Given observation data \(X\) and \(Y\), where \(y_i = T(x_i)\) and a basis set of functions or observables
one can define the transformed data \(\Psi_X\) and \(\Psi_Y\) with
yielding
Here, \(K^\top\in\mathbb{R}^{k\times k}\) acting from the left is the projection of the true Koopman operator \(\mathcal{K}\) into the basis \(B\).
Usually one performs rank-truncation or has over-/underdetermined systems. In such cases, \(K\) is determined via the pseudo-inverse \(K=\Psi_Y\Psi_X^\dagger\).
In case of a basis \(B\) which just contains the identity, EDMD reduces back to DMD.
To demonstrate EDMD and deeptime’s API, we consider a two-dimensional triple-well potential [2]. It is defined on \([-2,2]\times [-1, 2]\) and looks like the following:
[1]:
import numpy as np
import matplotlib.pyplot as plt
from deeptime.data import triple_well_2d
system = triple_well_2d()
x = np.linspace(-2, 2, num=100)
y = np.linspace(-1, 2, num=100)
XX, YY = np.meshgrid(x, y)
coords = np.dstack((XX, YY)).reshape(-1, 2)
V = system.potential(coords).reshape(XX.shape)
plt.contourf(x, y, V, levels=np.linspace(-4.5, 4.5, 20), cmap='coolwarm');
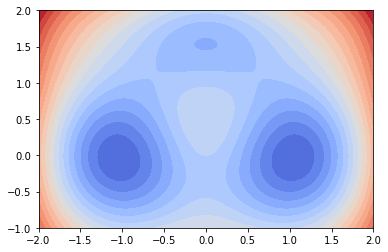
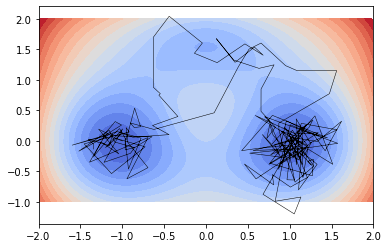
Inside this potential landscape we integrate particles subject to the stochastic differential equation
with \(\sigma=1.09\), using an Euler-Maruyama integrator.
[2]:
traj = system.trajectory(x0=[[-1, 0]], length=200, seed=66)
plt.contourf(x, y, V, levels=np.linspace(-4.5, 4.5, 20), cmap='coolwarm')
plt.plot(*traj.T, c='black', lw=.5);
To approximate the Koopman operator, we first select 25000 random points inside the domain and integrate them for 10000 steps under an integration step of \(h=10^{-5}\):
[3]:
N = 25000
state = np.random.RandomState(seed=42)
X = np.stack([state.uniform(-2, 2, size=(N,)), state.uniform(-1, 2, size=(N,))]).T
Y = system(X, n_jobs=8)
Now we pick a basis \(B\) to be all monomials up to and including degree \(\mathrm{deg}=10\).
[4]:
from deeptime.basis import Monomials
basis = Monomials(p=10, d=2)
Using this basis, we can fit an EDMD estimator and obtain the corresponding model.
[5]:
from deeptime.decomposition import EDMD
edmd_estimator = EDMD(basis, n_eigs=8)
edmd_model = edmd_estimator.fit((X, Y)).fetch_model()
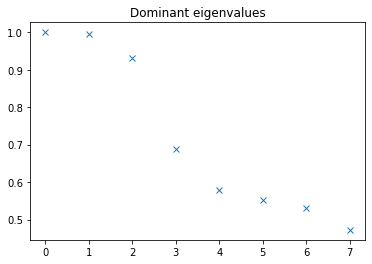
We can obtain the dominant eigenvalues associated to processes in the system.
[6]:
plt.plot(np.real(edmd_model.eigenvalues), 'x')
plt.title('Dominant eigenvalues');
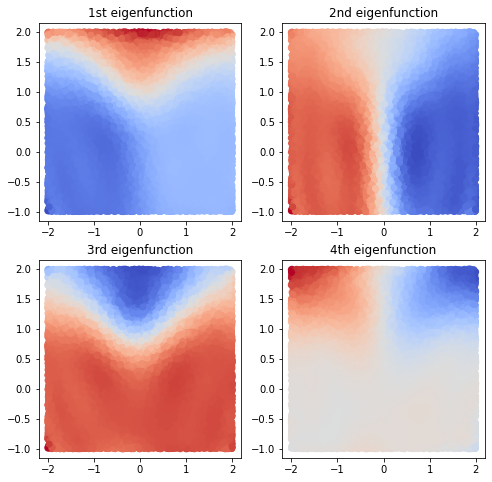
We project our input data \(X\) using the eigenfunctions:
[7]:
phi = np.real(edmd_model.transform(X, propagate=False))
# normalize
for i in range(len(edmd_model.eigenvalues)):
phi[:, i] = phi[:, i] / np.max(np.abs(phi[:, i]))
And visualize the first four eigenfunctions. They contain information about metastable regions and slow transitions.
[8]:
fig = plt.figure(figsize=(8, 8))
gs = fig.add_gridspec(ncols=2, nrows=2)
ax = fig.add_subplot(gs[0, 0])
ax.scatter(*X.T, c=phi[:, 0], cmap='coolwarm')
ax.set_title('1st eigenfunction')
ax = fig.add_subplot(gs[0, 1])
ax.scatter(*X.T, c=phi[:, 1], cmap='coolwarm')
ax.set_title('2nd eigenfunction')
ax = fig.add_subplot(gs[1, 0])
ax.scatter(*X.T, c=phi[:, 2], cmap='coolwarm')
ax.set_title('3rd eigenfunction')
ax = fig.add_subplot(gs[1, 1])
ax.scatter(*X.T, c=phi[:, 3], cmap='coolwarm')
ax.set_title('4th eigenfunction');
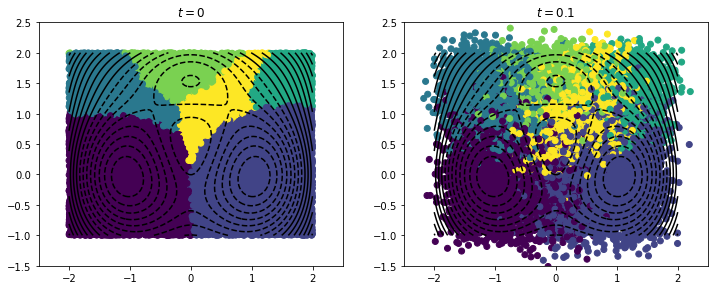
Using a clustering can reveal temporally coherent structures.
[9]:
from deeptime.clustering import KMeans
kmeans = KMeans(n_clusters=6, n_jobs=4).fit(np.real(phi)).fetch_model()
c = kmeans.transform(np.real(phi))
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
ax1.scatter(*X.T, c=c)
ax1.set_title(r"$t = 0$")
ax1.set_aspect('equal')
ax1.set_xlim([-2.5, 2.5])
ax1.set_ylim([-1.5, 2.5])
ax1.contour(x, y, V, levels=np.linspace(-4.5, 4.5, 20), colors='black')
ax2.scatter(*Y.T, c=c)
ax2.set_title(r"$t = 0.1$")
ax2.set_aspect('equal')
ax2.set_xlim([-2.5, 2.5])
ax2.set_ylim([-1.5, 2.5])
ax2.contour(x, y, V, levels=np.linspace(-4.5, 4.5, 20), colors='black');
References
